Hanxue Gu
Welcome to Hanxue's homepage!

hanxue (dot) gu (at) duke (dot) edu
Duke University
Durham, NC, United States, 27703
About me: I am a 5th year Ph.D. student in Electrical and Computer Engineering at Duke University, working at the intersection of AI and Healthcare. I am fortunate to be advised by Prof. Maciej A. Mazurowski under Duke Spark Initiative. My research sits at the intersection of machine learning and healthcare, with a focus on developing and adapting deep learning methods for medical image analysis—from application-oriented tools to foundational advancements.
I am interested in several research topics, including foundation models, self- and semi-supervised learning, image harmonization, and clinically meaningful AI. My work spans segmentation, registration, 2D-to-3D reconstruction, and risk prediction. See my Research Topics Bar for details.
Before starting my Ph.D., I received my Bachelor’s degree from Zhejiang University, where I studied Electrical & Information Engineering and minored in ACEE under the Chu Kochen Honors College. Prior to Duke, I spent a year as a research intern at the Athinoula A. Martinos Center for Biomedical Imaging at Massachusetts General Hospital (MGH), where I worked on optical image reconstruction and deep learning.
See my Google Scholar for a full list of my publications, with a few highlights shown below.
Feel free to connect with me on LinkedIn or contact me through email— I’m always happy to chat! (🤗)
news
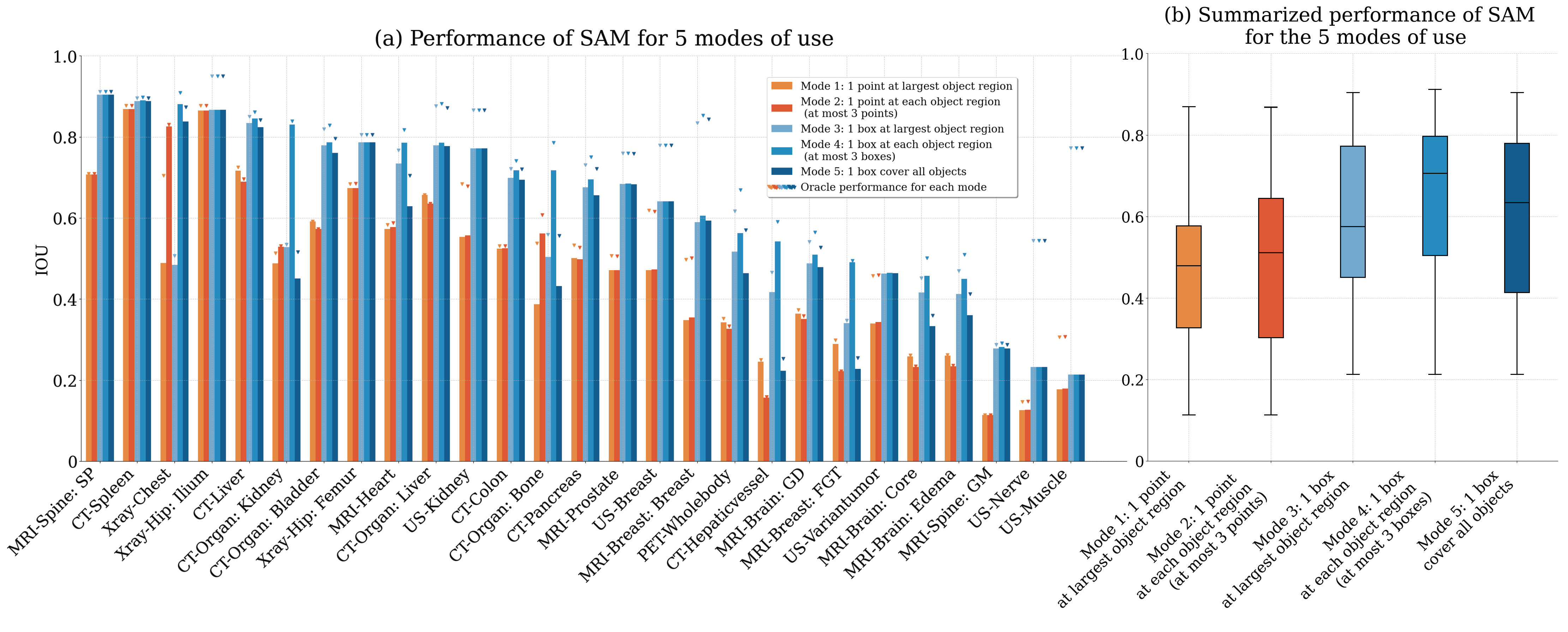
| Aug 02, 2025 | 🎉 Thrilled to share that two of our papers on breast MRI have been accepted to the Deep-Breath Workshop at MICCAI 2025! 🧠 The first paper presents the most comprehensive breast MRI segmentation dataset to date, covering a wide range of anatomical structures. We hope this effort can lay the foundation for future research in breast composition analysis, risk stratification, and AI-driven screening tools. 🌀 The second paper focuses on generalizable breast MRI registration, addressing the challenge of aligning highly deformable anatomy across patients and imaging sessions. We demonstrate the power of foundation model features and training-free registration strategies to enable robust alignment without manual annotation. We believe these works together push toward more generalizable, scalable, and interpretable tools for breast MRI analysis. 🚀 Stay tuned — datasets, code, and more resources will be released soon. Big thanks to the Deep-Breath organizers and all our co-authors and collaborators! |
|---|---|
| Jun 29, 2025 | 🩺 Excited to share that our paper “Breast density in MRI: an AI-based quantification and relationship to assessment in mammography” has been accepted for publication in npj Breast Cancer! In this study, we developed an AI-driven pipeline to quantify breast density directly from MRI scans and explored how these MRI-derived metrics relate to traditional density assessments from mammography. Our work provides new insights into leveraging MRI for more comprehensive breast density evaluation. Huge thanks to all co-authors and collaborators — looking forward to seeing this work published soon! |
| Jun 29, 2025 | 🎉 Excited to share that the paper I contributed as co-author “SAMora: Enhancing SAM through Hierarchical Self-Supervised Pre-Training for Medical Images” has been accepted to ICCV 2025! In this work, we propose SAMora, a novel framework that boosts the performance of the Segment Anything Model (SAM) on medical images through a hierarchical self-supervised pre-training strategy. Our approach brings significant improvements to medical segmentation tasks where labeled data is scarce. Thank you to all co-authors and collaborators for making this possible! |
| Jun 25, 2025 | 🧠 Excited to share our latest work “MRI-CORE: A Foundation Model for Magnetic Resonance Imaging”, now available on arXiv! This project introduces a general-purpose MRI foundation model capable of handling diverse MRI sequences and anatomical regions, laying the groundwork for improved downstream tasks across medical imaging applications. 📄 Read the paper on arXiv Thank you to all collaborators for making this happen! |
| Jun 12, 2025 | 🎉 Our paper “Deep Learning Automates Cobb Angle Measurement Compared with Multi-Expert Observers” has been accepted to the British Journal of Radiology: Artificial Intelligence! This work presents a clinically interpretable AI model for scoliosis assessment, validated in a multi-reader study against experienced clinicians. Big thanks to our collaborators! |
selected publications
-
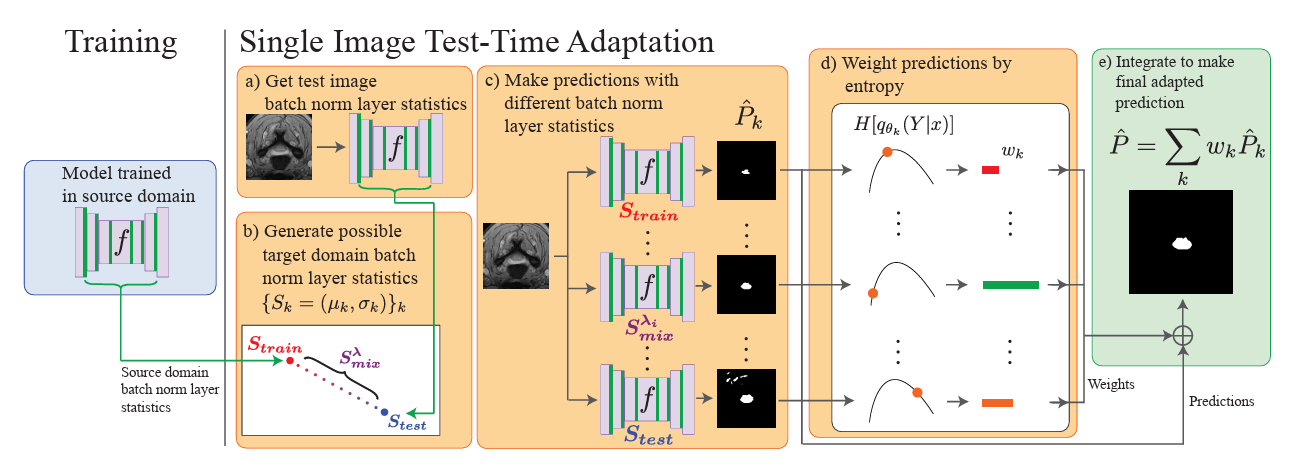 Medical image segmentation with intent: Integrated entropy weighting for single image test-time adaptationIn Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, May 2024
Medical image segmentation with intent: Integrated entropy weighting for single image test-time adaptationIn Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, May 2024